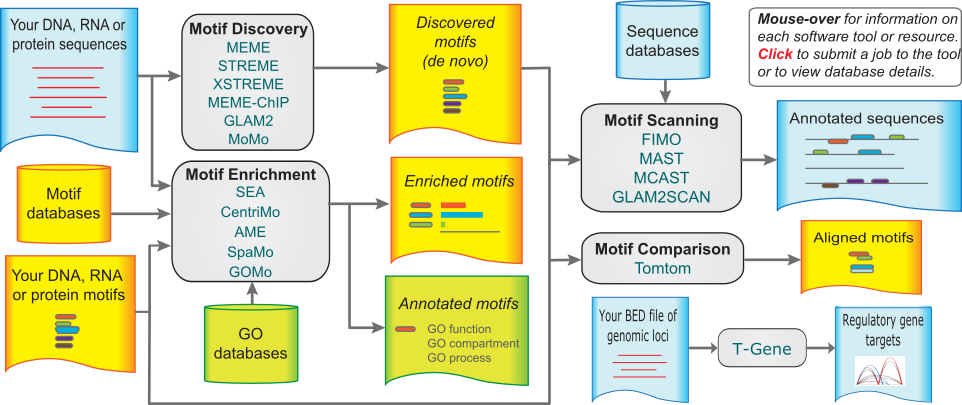
The MEME Suite
Motif-based sequence analysis tools
Development of the MEME Suite was funded by grant R01 GM103544 from the National Institutes of Health.
| Developed and maintained by | |||
|---|---|---|---|

|

|

|
|

Development of the MEME Suite was funded by grant R01 GM103544 from the National Institutes of Health.
| Developed and maintained by | |||
|---|---|---|---|

|

|

|
|